Microbial Processes in Anoxic Ecosystems

We use environmental metagenomics, laboratory cultivation and microcosms to uncover the microbial communities in anoxic ecosystems. A particular focus is on microbial communities in peatlands to understand the microbial processes in naturally pristine sites, and restorability of sites that have been impacted by human activities.
Projects:
Microbial Processes Influencing Methane Cycling in Pristine Northern Peatlands
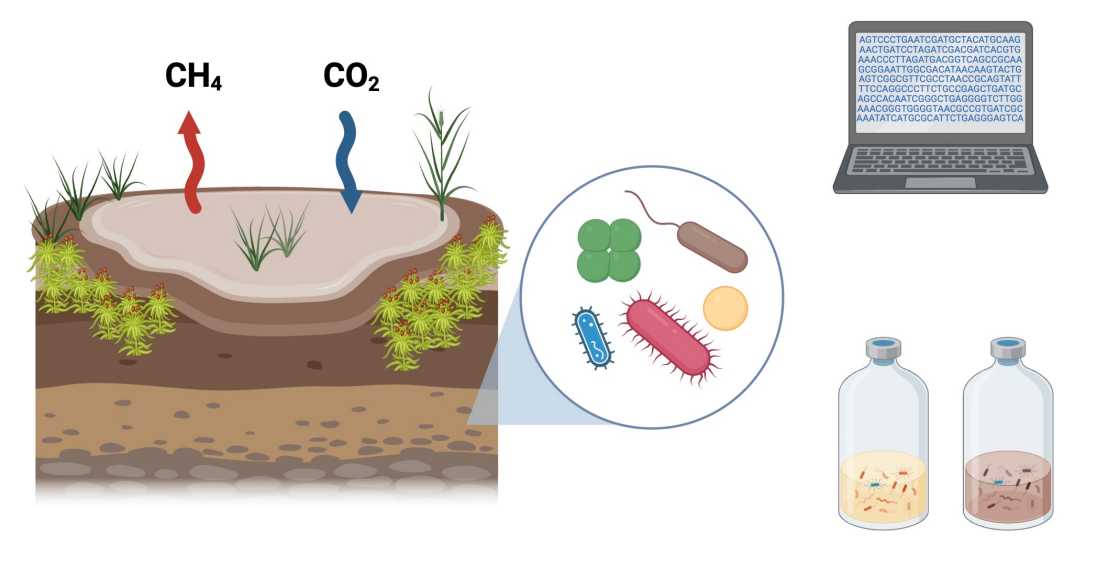
We investigate the microbial processes driving methane (CH₄) cycling in pristine northern peatlands, correlating them with the biogeochemistry of the sites. Through metagenomics and metatranscriptomics, we explore their metabolic potential and activity, while identifying extrachromosomal DNA (ecDNA) elements that may shape CH₄ turnover in soil. Additionally, we establish anaerobic enrichment cultures targeting one-carbon (C1)-metabolizing microorganisms, further examining their metabolism and characterizing associated ecDNA elements that could drive CH₄ cycling.
Project leader: Giulia Fiorito
Microbial Processes in Swiss Peatlands
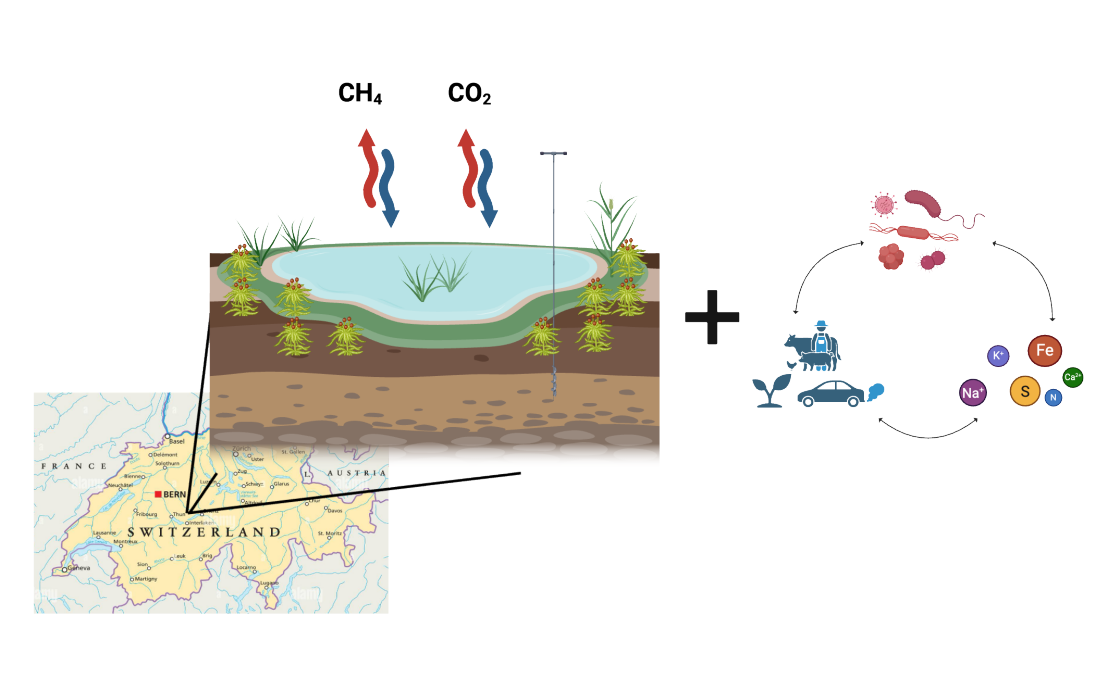
Peatlands cover just 3% of the Earth's land surface but store more than a third of the total soil organic carbon. However, the impact of climate change on microbial communities and carbon cycling in these ecosystems remains unclear. Our research integrates fieldwork, laboratory experiments, and computational approaches to uncover how microbial communities influence biogeochemical processes in peatlands.
Project leader: Madison Barney
WLP project

We investigate the global distribution of extrachromosomal elements and microbial genomes harboring Wood-Ljungdahl Pathway (WLP) genes. Our research explores how extrachromosomal elements influence microbial metabolism and ecological interactions, revealing their previously overlooked role in carbon cycling and energy conservation.
Lake Cadagno

We are investigating microbial communities and
their activity in the anoxic sediments of meromictic Lake Cadagno. Our
focus is on microbes involved in the carbon and sulfur cycle, aiming to
understand their contribution to geochemical cycles.
Project leader: Bledina Dede
Nanopore sequencing for decoding epigenetic patterns in natural microbial communities

Using Nanopore sequencing of wetland microbial communities, we investigate DNA modifications to reveal microbial epigenetic diversity and leverage epigenetic patterns to improve genome binning and host-extrachromosomal element linkage.
Project leader: Jo Herbert